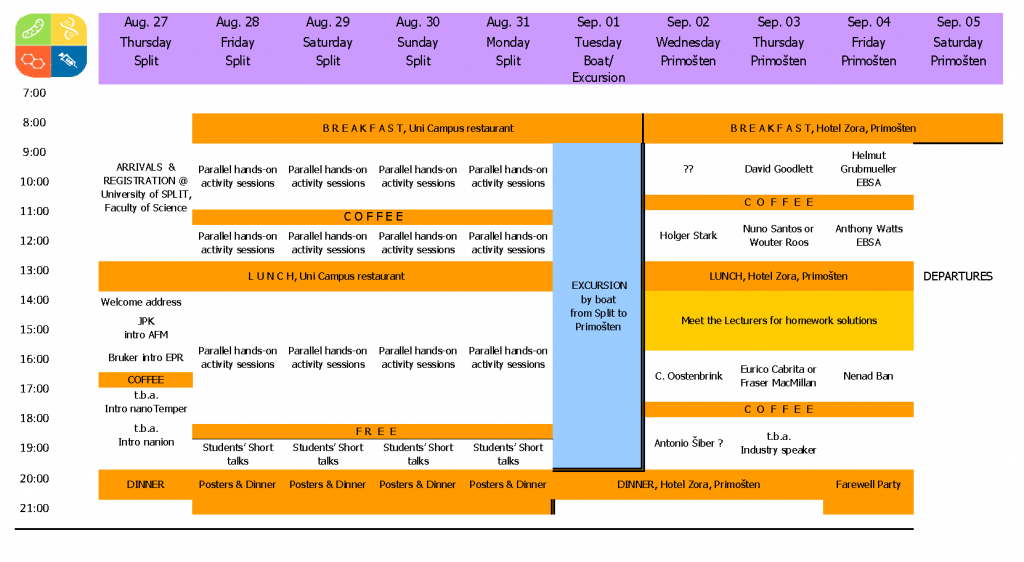
Program
EBSA key lecturers:
- Nenad Ban, ETH, Zürich, Switzerland: Protein structure& function; X-ray diffraction
- Ribosomes and their functional complexes
- Beyond the prokaryotic ribosome
- Helmut Grubmüller, Universität Göttingen, Germany: Protein structure& function; cryoEM
- Theoretical and Computational Biophysics
- Anthony Watts, Department of Biochemistry, Oxford, UK: Protein structure& function; solid state NMR
- Principles of solid state NMR for the study of biomolecules
- Solid state NMR for structural studies of large integral membrane proteins
- Frances Separovic, Bio21 Institute, Uni Melbourne, Australia: Protein structure& function; solid state NMR
Special Guest Lecturer:
Theoretical simulations:
- Chris Oostenbrink, UNRLF, Vienna, Austria: Structure and dynamics of biomolecules; modeling
- Ensembles and sampling, leading to molecular dynamics simulations
- Structure refinement using molecular dynamics simulations (NMR observables)
- Calculation of free energies from molecular simulation
- Antonio Šiber, Institute of physics, Zagreb, Croatia: Physics of viruses
- Physics of viruses: electrostatics, elasticity and DNA condensation in viruses 1,2,3
- Lynn Kamerlin, Uppsala University, Sweden: Simulating large scale conformational changes in biomolecular systems
- Introduction to a sampling molecular dynamics simulation technique known as Hamiltonian replica exchange (HREX)
Molecular Biophysics:
Programme & organization Committee
- Mario Cindrić, Ruđer Bošković Institute, Zagreb, Croatia
- Ida Delač Marion, Institute of Physics, Zagreb, Croatia
- Amela Hozić, Ruđer Bošković Institute, Zagreb, Croatia
- Nadica Ivošević DeNardis, Ruđer Bošković Institute, Zagreb, Croatia
- Lucija Krce, University of Split, Croatia
- Nadica Maltar-Strmečki, Ruđer Bošković Institute, Zagreb, Croatia
- Antonio Šiber, Institute of Physics, Zagreb, Croatia
- Tomislav Vuletić, Institute of physics, Zagreb, Croatia (chair)
- Larisa Zoranić, University of Split, Croatia

Top


