Program
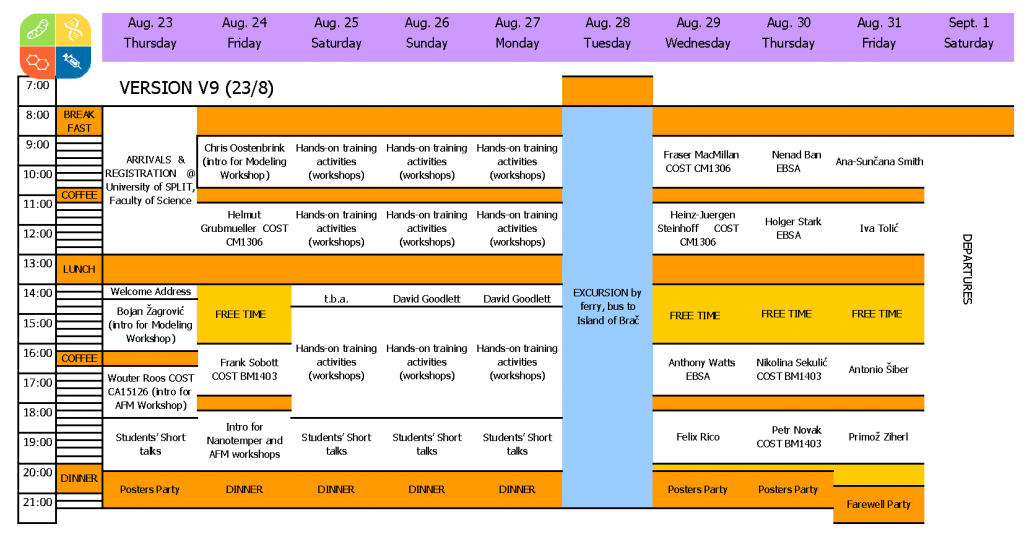
EBSA key lecturers:
- Nenad Ban, ETH, Zürich, Switzerland: Protein structure& function; X-ray diffraction
- Ribosomes and their functional complexes
- Beyond the prokaryotic ribosome
- Holger Stark, MPI-BPC, Göttingen, Germany: Protein structure& function; cryoEM
- Introduction into single particle cryo-EM
- High-resolution structure determination of macromolecular complexes by cryo-EM
- Anthony Watts, Department of Biochemistry, Oxford, UK: Protein structure& function; solid state NMR
- Principles of solid state NMR for the study of biomolecules
- Solid state NMR for structural studies of large integral membrane proteins
- Receptor dynamics and structure in membranes resolved using solid state NMR
Special Guest Lecturers:
- David Goodlett, School of Pharmacy, University of Maryland
- Felix Rico, FM4B-Lab, U1006 INSERM & Aix-Marseille Université, Marseille, France
- Mechanics of single biomolecules probed by atomic force microscopy
Physics of cells by Croatia ERC grantees:
- Ana-Sunčana Smith, Ruđer Bošković Institute, Zagreb, Croatia
- Membranes, from self assembly to rafts 1,2,3
- Iva Tolić, Ruđer Bošković Institute, Zagreb, Croatia: Physics of cells
- Forces in the mitotic spindle
- Live-cell fluorescence microscopy and laser-cutting inside the cell
Theoretical simulations:
- Chris Oostenbrink, UNRLF, Vienna, Austria: Structure and dynamics of biomolecules; modeling
- Ensembles and sampling, leading to molecular dynamics simulations
- Structure refinement using molecular dynamics simulations (NMR observables)
- Calculation of free energies from molecular simulation
- Antonio Šiber, Institute of physics, Zagreb, Croatia: Physics of viruses
- Physics of viruses: electrostatics, elasticity and DNA condensation in viruses 1,2,3
- Primož Ziherl, JSI, Ljubljana, Slovenia
- Bojan Žagrović, MFPL, Vienna, Austria: Computational Biophysics of Macromolecules
- More dynamic than we think? On conformational averaging in structural biology 1, 2
- Protein-RNA interactions and the origin of the genetic code
Between Atom and Cell (COST Action CA15126 ARBRE MOBIEU)
- Nadica Ivošević DeNardis, Ruđer Bošković Institute, Zagreb, Croatia
- Tea Mišić Radić, Ruđer Bošković Institute, Zagreb, Croatia
- Tomasz Kobiela, Institute of Biotechnology, Warsaw University of Technology, Poland
- Tomislav Vuletić, Institute of physics, Zagreb, Croatia
Native Mass Spectrometry (COST Action BM1403):
- Mario Cindrić, Ruđer Bošković Institute, Zagreb, Croatia
- Mass Spectrometry for protein sequencing
- Amela Hozić, Ruđer Bošković Institute, Zagreb, Croatia
- Frank Sobott, Uni Antwerpen, Belgium
- “native” mass spectrometry, ion mobility: structural proteomics techniques
- Nikolina Sekulić, Centre for Molecular Medicine, University of Oslo, Norway
- Petr Novák, BIOCEV, Biotechnology and Biomedicine Center of the Academy of Sciences and Charles University in Vestec, Czech Republic
Molecular Machines (COST Action CM1306):
Programme & organization Committee
- Mario Cindrić, Ruđer Bošković Institute, Zagreb, Croatia
- Ida Delač Marion, Institute of Physics, Zagreb, Croatia
- Amela Hozić, Ruđer Bošković Institute, Zagreb, Croatia
- Nadica Ivošević DeNardis, Ruđer Bošković Institute, Zagreb, Croatia
- Lucija Krce, University of Split, Croatia
- Nadica Maltar-Strmečki, Ruđer Bošković Institute, Zagreb, Croatia
- Tea Mišić Radić, Ruđer Bošković Institute, Zagreb, Croatia
- Martina Požar, University of Split, Croatia
- Antonio Šiber, Institute of Physics, Zagreb, Croatia
- Tomislav Vuletić, Institute of physics, Zagreb, Croatia (chair)
- Larisa Zoranić, University of Split, Croatia

Top


